
Chromosome Organization
Chromosomes carry the information to generate a living cell. In both prokaryotes and eukaryotes, chromosomal DNA is highly compacted to fit inside its cellular confinement. This implies a major organizational problem: the DNA does not only have to be highly condensed, but its spatial organization also has to facilitate processes such as transcription and replication. However, it’s still an outstanding puzzle how organized the chromosomes really are, and how this organization affects their functionality.
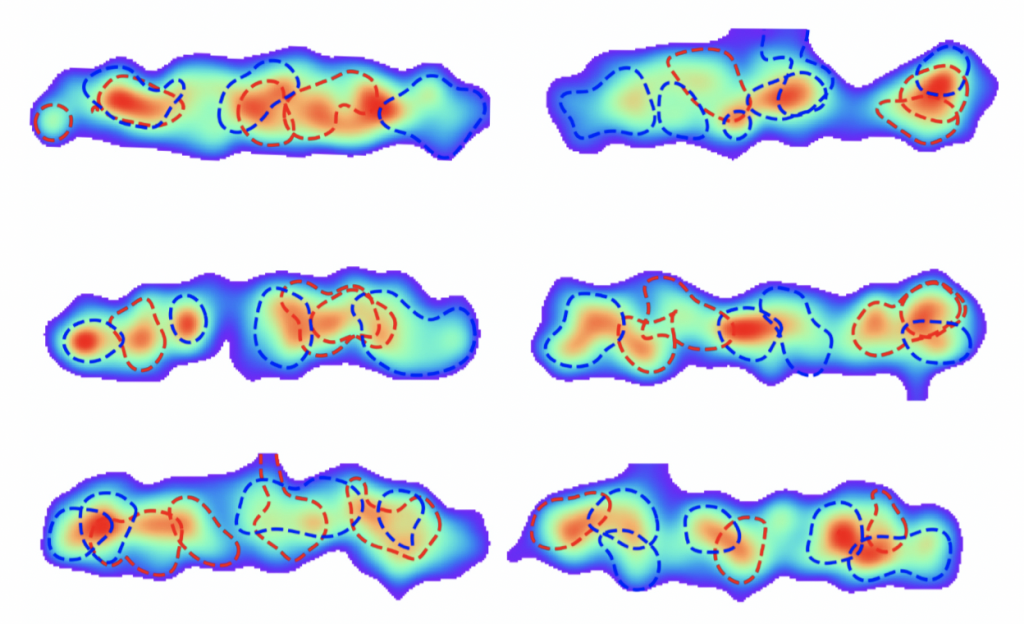
We are intrigued by the degree of chromosomal organization in bacterial cells. The classical picture of an amorphous bacterial chromosome has gradually been broken down, thanks to experimental techniques such as fluorescence microscopy and Hi-C. These Hi-C experiments measure pair-wise contacts between chromosomal loci averaged over a population of cells, providing key data on spatial chromosome organization. Despite all experimental progress, however, the organization and variability of the bacterial chromosome across genomic scales remain unclear. It is still a major outstanding challenge to interpret such Hi-C data. We address this outstanding problem, by developing a principled and fully-data driven Maximum Entropy approaches to faithfully extract the distribution of single-cell 3D chromosome conformations from Hi-C data. This allows us to elucidate the full spatial organization of the bacterial chromosome across genomic scales.

The bacterial chromosome is organized in a structure called the nucleoid in part by an array of so-called Nucleoid-Associated Proteins (NAPs). Such NAPs play a role in DNA processes such as transcription, recombination, gene regulation, and DNA segregation. The biological function of these NAPs relies on them binding to DNA in large numbers. Collectively, DNA-bound NAPs can control chromosome structure by interacting with each other and with DNA. Experiments show that NAPs can mechanically interact with the DNA, inducing bending, twisting, looping, or coiling of the DNA. Some NAPs are thought to form large protein droplets on the DNA, while others act as molecular motors to actively extrude DNA loops. The ability of NAPs to locally structure DNA has been found to play a role in large-scale DNA organization. Our goal is to elucidate the physical principles underlying the origin and function of the organization of the bacterial chromosome by such Nucleoid-Associated Proteins.
Further reading:
Resolving the degree of order in the bacterial chromosome using a statistical physics approach
Joris J. B. Messelink, Jacqueline Janssen, Muriel C. F. van Teeseling, Martin Thanbichler, Chase P. Broedersz
Nature Communications (in press) [arXiv:2002.03880]
Theory of Active Intracellular Transport by DNA-relaying
Christian Hanauer, Silke Bergeler, Erwin Frey, Chase P. Broedersz
submitted for publication [arXiv:2101.02640]
Bacterial chromosome organization by collective dynamics of SMC condensins
Christiaan A. Miermans, Chase P. Broedersz
J. R. Soc. Interface 15, 20180495 (2018) [arXiv:1805.03983]
Looping and clustering model for the organization of partitioning proteins on the bacterial genome
Jean-Charles Walter, Jérôme Dorignac, Frédéric Geniet, John Palmeri, Andrea Parmeggiani, Ned S. Wingreen, Chase P. Broedersz
New J. Phys. 20, 035002 (2018)[arXiv:1707.01373]
Condensation and localization of the partitioning protein ParB on the bacterial chromosome
Chase P. Broedersz, Xindan Wang, Yigal Meir, Joseph J. Loparo, David Z. Rudner, Ned S. Wingreen
PNAS 111 (24), 8809-8814 (2014)